What do specific lipids "do" in our brains?
Lipids are often stigmatized as boring or dangerous structural macromolecules compared to proteins and nucleic acids. However, lipids can signal dynamically and also pervasively control protein localization and function by compartmentalizing biochemical reactions. Lipid diversity and asymmetry define the cell as we understand it, and lipid dysregulation causes diseases. However, our ability to manipulate lipids remains poor. Our lab uses Drosophila genetics to predictably and precisely alter brain lipids to decipher lipid functions. How is the brain’s lipidome established and maintained for life across diverse neurons and glia? Why are certain lipids deployed by specific cells and compartments, ie synaptic vesicles? Can better quantitation and manipulation of brain lipids using novel genetic and mass-spectrometry tools reveal underlying principles of metabolic networks that are dysregulated in aging and disease?
Questions we ask
How do brains build And Maintain distinct Cellular lipidomes?
|
The brain is composed of neurons and glia, which largely perdure for life despite high membrane flux at synapses. We analyze brain lipids from controls and genetic mutants to understand how the brain is built and maintained, and how cell-specific gene regulatory networks encode tissue and cellular lipidomes.
|
FINDING FUNCTIONS FOR LIPIDS
|
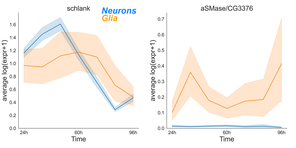
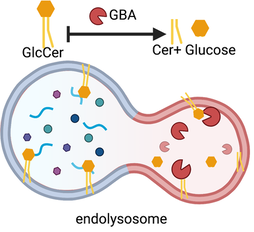
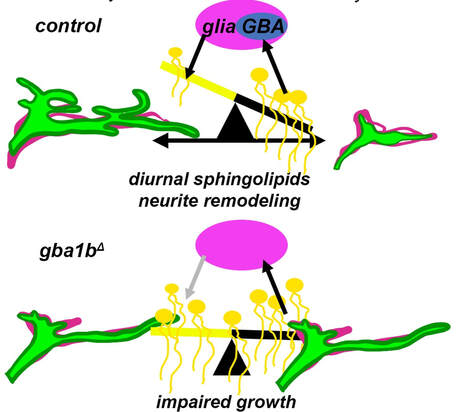
We found that sphingolipids sculpted neural circuit remodeling, with inputs from neural biosynthesis and glial catabolism in lysosomes.
What do sphingolipids and other lipids do across neural and glial compartments, including in lysosomes and synaptic vesicles? We will identify functions for brain lipids by precise genetic interventions, quantitative lipidomics, and cellular and behavioral phenotyping to link specific lipid molecules to cellular functions. |
HOW DO LIPID Alterations Foment AGING AND DISEASE?
Why do specific cells suffer in particular diseases? Could an underlying lipid code dictate cellular sensitivity to aging and degeneration? Do certain defects arise during development when lipid flux is highest? We seek to quantitate and manipulate the aging lipidome with unbiased genetic and dietary approaches, as well as those targeted to known risk factors for neurodegenerative diseases like Parkinson’s and Alzheimer’s
Tools we use
drosophila genetics
|
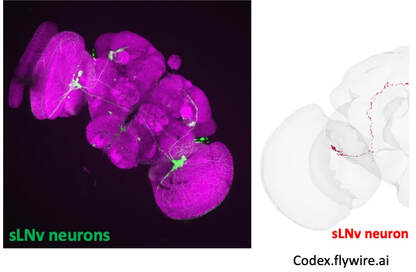
Drosophila neurobiologists live blissfully in a golden era, with genetic access to nearly every cell type in the brain, a complete electron microscope connectome, and libraries of RNAi and mutants. Overexpression and knockout/knockdown approaches have already yielded rich insight into how certain genes control the brain's lipid profile.
However, the reagents that have worked historically for analyzing protein gain and loss of function are less amenable to lipid enzyme perturbation, ie typical overexpression can drive lethality as cells are sensitive to the precise level of lipid-regulating enzymes and the lipids they produce. Moreover, genetic interference with cellular lipidomes will undoubtedly trigger rapid compensatory mechanisms, meaning that a goal of the lab is to achieve more acute spatiotemporal lipidome manipulations in defined cell types before phenotyping. We are eager to build better tools! |
CONFOCAL MICROSCOPY
|
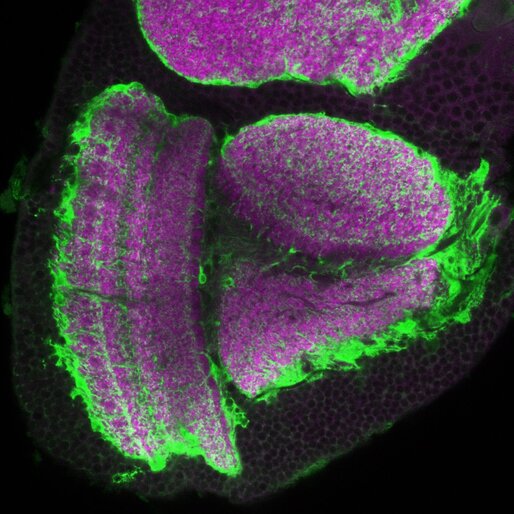
Imaging is one of the joys of a cell and developmental biologist! We can peer inside the brain at cell morphology, fate, wiring, and even physiology with 2photon live imaging technologies. A lab staple is confocal light microscopy to detect specific antibodies or live stains. On the right is a janky video of imaging for protein ubiquitin aggregates (red) and a neural CD8::GFP marker that is particularly enriched in the mushroom bodies.
In the future, we hope to incorporate genetically-encoded lipid dyes and live sensors of membrane order such as Laurdan. |
|
LIQUID CHROMATOGRAPHY-MASS SPECTROMETRY (LC-MS)
|
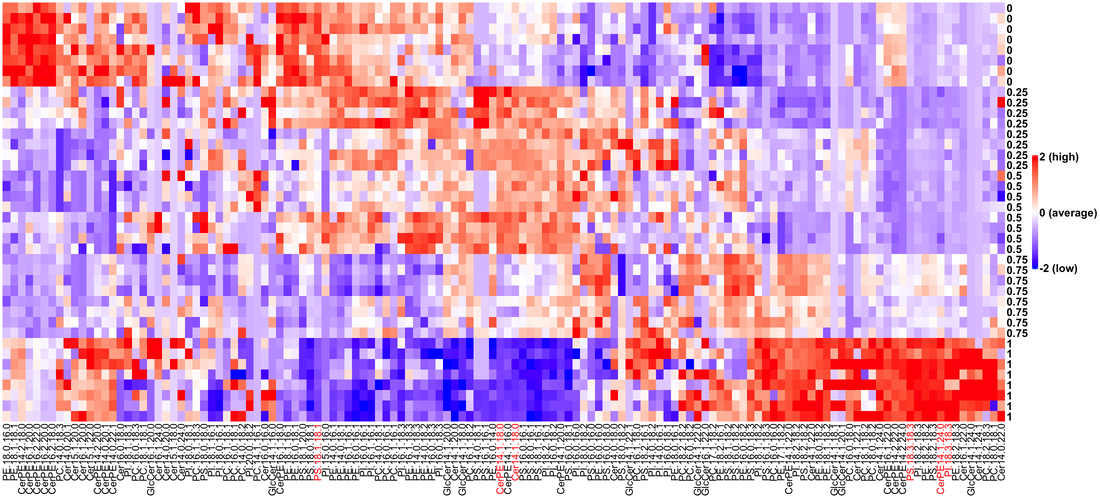
We microdissect brain tissue for lipidomics (and can further purify cells or compartments by FACS or standard biochemical techniques). Brains are snap-frozen, polar lipids extracted (Folch), and then analyzed by LC-MS using an Agilent triple quadrupole with our collaborators as well as the UCSF QMAC core. We are actively working on genetic strategies to improve cell-type specific lipidomic resolution by lipid tagging.
We are also exploring untargetted shotgun lipidomics in parallel to targeted, but vast improvements are needed in library matching to reliably identify the full scope of the unknown fragments. The right video shows a tube of dissected brains in methanol before placement on dry ice. Time is of the essence to prevent lipid peroxidation! |
|
Computation
|
Lipidomics and RNA-seq datasets yield large datasets that require computational analyses for visualization and understanding.
Moreover, with the advent of alphafold, we can model proteins and protein-membrane interactions in simplified membranes. We are hoping to collaborate with biophysicists to model mutant and wild-type brain membrane composition, and its effects of neural function in EM-defined cellular morphologies. |
BEHAVIOR
|
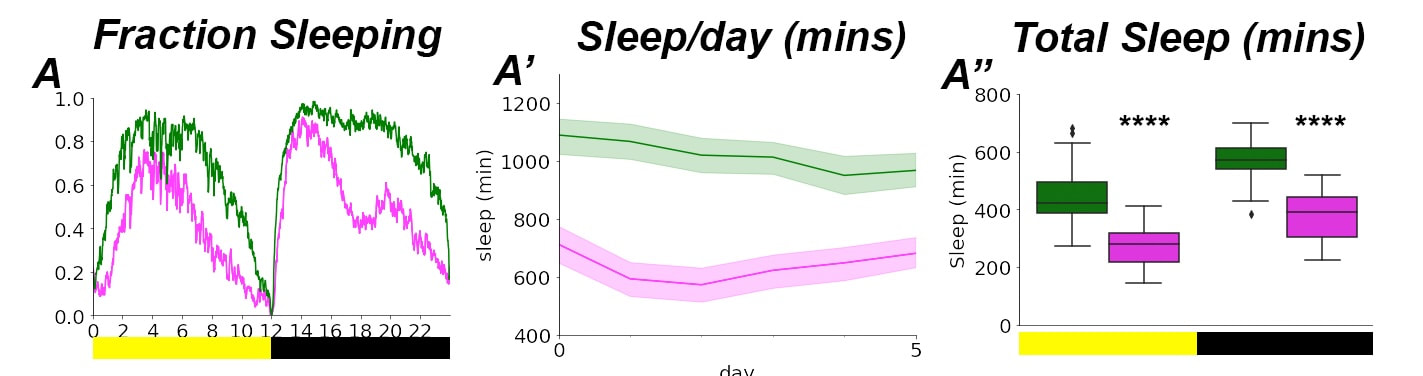
Along with possessing brains, flies also sleep and have circadian rhythms, with peaks of activity and dawn and dusk. We can quantify circadian behavior and sleep using activity monitors. To the right, controls (green) versus sphingolipid mutant (magenta) flies show different sleep patterns. Disrupted circadian rhythm often precedes neurodegenartion, and sleep is considered neuroprotective. Much work remains to decipher the complex relationship between diet, sleep, and brain health.
|